
new engines in maize
extending_kernlab_support.Rmd
library(ggplot2)
library(parsnip)
library(maize)
#>
#> Attaching package: 'maize'
#> The following object is masked from 'package:parsnip':
#>
#> check_args
set.seed(31415)
corn_train <- corn_data |> dplyr::sample_frac(.80)
corn_test <- corn_data |> dplyr::anti_join(corn_train)
#> Joining with `by = join_by(height, kernel_size, type)`
# use this later to create a classification field
# -----------------------------------------------
kernel_min <- corn_data$kernel_size |> min()
kernel_max <- corn_data$kernel_size |> max()
kernel_vec <- seq(kernel_min, kernel_max, by = 1)
height_min <- corn_data$height |> min()
height_max <- corn_data$height |> max()
height_vec <- seq(height_min, height_max, by = 1)
corn_grid <- expand.grid(kernel_size = kernel_vec, height = height_vec)
# -----------------------------------------------kernels for the ksvm engine
{maize} is continuing down the path of least known engines, novelties, and unique ML methods.
Initially, {maize} was created to add additional kernel support to {parsnip}, outside of the known linear, polynomial, and radial basis function kernels. This added a handful of very odd kernels from within the {kernlab} library. After this first leap, there were a few customized kernels added and registered within {kernlab} and {maize}, based on other efforts by Python and Julia open sourced packages.
SVM
the main engine supported within {maize} is
kernlab::ksvm(). This function gave a lot of flexibility
around extending kernels within the package. A typical regression model
fit would be like so:
# model params --
svm_reg_spec <-
svm_laplace(cost = 10, margin = .01) |>
set_mode("regression") |>
set_engine("kernlab")
# fit --
svm_reg_fit <- svm_reg_spec |> fit(height ~ ., data = corn_train)
# predictions --
preds <- predict(svm_reg_fit, corn_test)
# plot --
corn_test |>
cbind(preds) |>
ggplot() +
geom_point(aes(x = kernel_size, y = height, color = "truth", shape = type), size = 3) +
geom_point(aes(x = kernel_size, y = .pred, color = "pred", shape = type), size = 3, alpha = .5) +
theme_minimal() +
labs(title = 'Laplacian Kernel SVM',
subtitle = "corn height prediction") 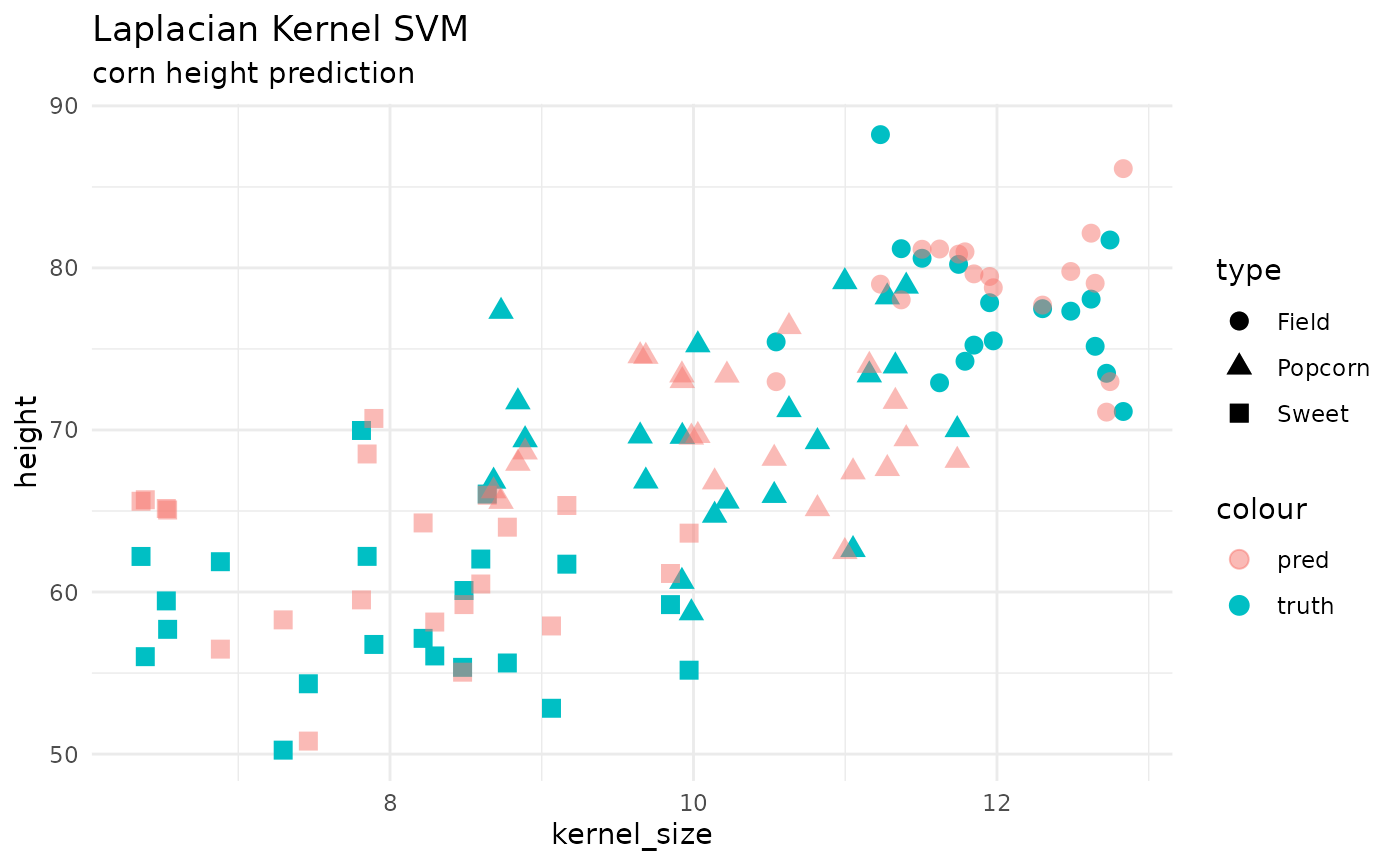
From there, the package started cooking up more stuff, like adding a few {recipes} related to kernel principal component analysis and Hebbian algorithms, which added new step_ functions to the package. Then in recent developments, post-processors were added for point and interval calibration.
beyond the ksvm engine
While the package is built around supporting kernels and, well, support vectors, there are many other engines that will be added to {maize}. There is a handful within the {kernlab} library, given we have already wrapped the “ksvm”, the next step would be adding some of the other engines within {kernlab}.
{maize} will also support {ebmc} methods for boosting and bagging SVMs from {e1701}. Another handful of engines come from {mildsvm} - shoutout to @jrosell for finding these. Support for these will be tested in the future.
KQR
kernlab::kqr() kernel quantile regression is new to
{maize}! below shows an example of how to use this engine for
regression. currently, the only kernel supported in
maize 0.0.1.9000 is “laplacian”.
# model params --
kqr_reg_spec_hi <-
kqr_laplace(cost = 10, tau = 0.95) |>
set_mode("regression") |>
set_engine("kernlab")
kqr_reg_spec_lo <-
kqr_laplace(cost = 10, tau = 0.05) |>
set_mode("regression") |>
set_engine("kernlab")
# fit --
kqr_reg_fit_hi <- kqr_reg_spec_hi |> fit(height ~ ., data = corn_train)
#> Using automatic sigma estimation (sigest) for RBF or laplace kernel
kqr_reg_fit_lo <- kqr_reg_spec_lo |> fit(height ~ ., data = corn_train)
#> Using automatic sigma estimation (sigest) for RBF or laplace kernel
# predictions --
preds_hi <- predict(kqr_reg_fit_hi, corn_test)
preds_lo <- predict(kqr_reg_fit_lo, corn_test)
# plot --
corn_test |>
cbind(hi = preds_hi$.pred) |>
cbind(lo = preds_lo$.pred) |>
ggplot() +
geom_point(aes(x = kernel_size, y = height, color = "truth", shape = type), size = 3) +
geom_point(aes(x = kernel_size, y = hi, color = "hi", shape = type), size = 3, alpha = .5) +
geom_point(aes(x = kernel_size, y = lo, color = "lo", shape = type), size = 3, alpha = .5) +
theme_minimal() +
labs(title = 'Laplacian Kernel KQR',
subtitle = "corn height prediction") 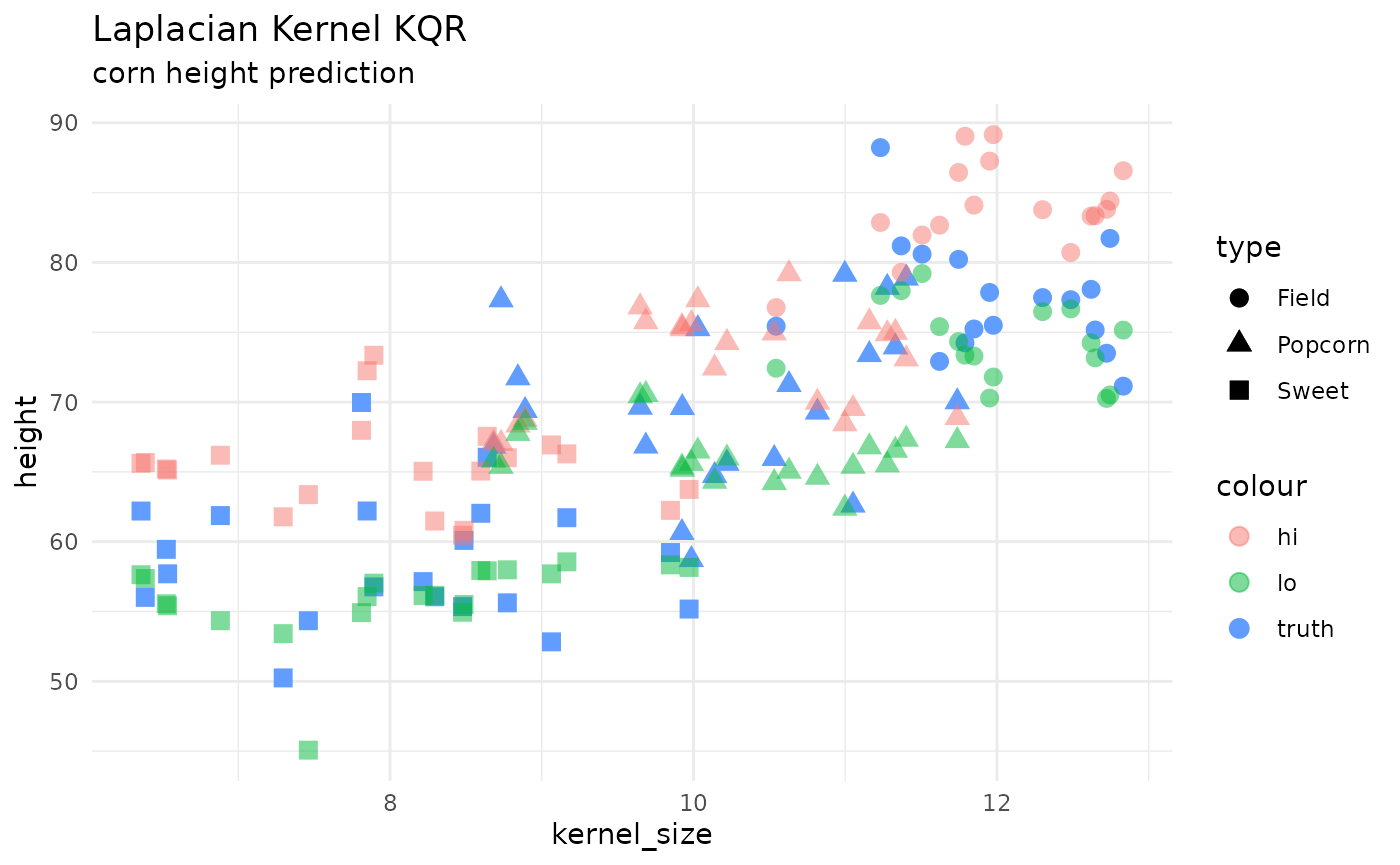
LSSVM
kernlab::lssvm() least squares support vector machine is
new to {maize}! below shows an example of how to use this engine for
classification. currently, the only kernel supported in
maize 0.0.1.9000 is “laplacian”.
# model params --
lssvm_cls_spec <-
lssvm_laplace(laplace_sigma = 20) |>
set_mode("classification") |>
set_engine("kernlab")
# fit --
lssvm_class_fit <- lssvm_cls_spec |> fit(type ~ ., data = corn_train)
# predictions --
# predictions --
preds <- predict(lssvm_class_fit, corn_grid, "class")
pred_grid <- corn_grid |> cbind(preds)
# plot --
corn_test |>
ggplot() +
geom_tile(inherit.aes = FALSE,
data = pred_grid,
aes(x = kernel_size, y = height, fill = .pred_class),
alpha = .8) +
geom_point(aes(x = kernel_size, y = height, color = type, shape = type), size = 3) +
theme_minimal() +
labs(title = "Laplacian Kernel LSSVM",
subtitle = "corn class prediction") +
scale_fill_viridis_d() +
scale_color_manual(values = c("violet", "cyan", "orange"))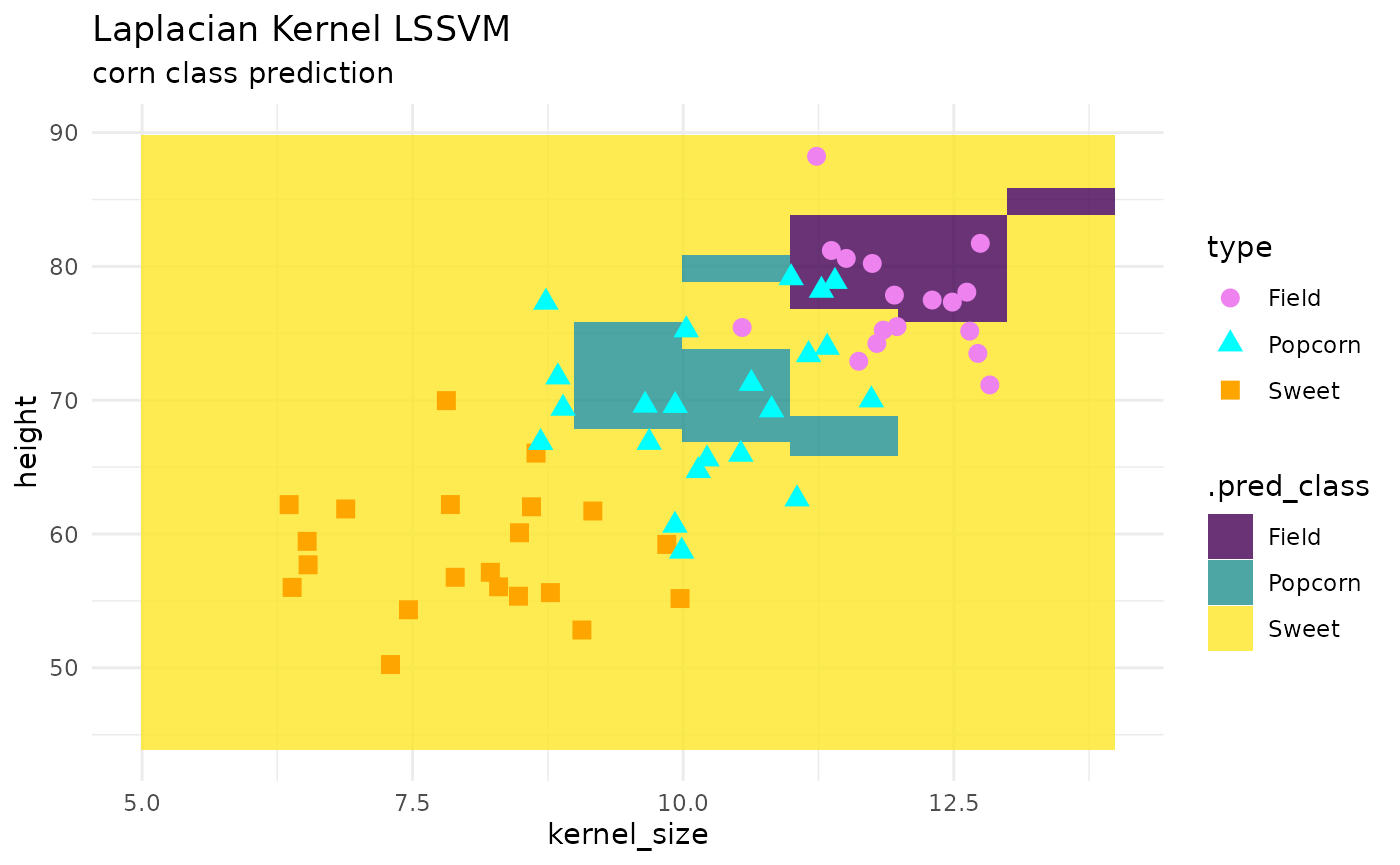
RVM
note the {kernlab} and {maize} implementations are for regression only.
kernlab::rvm() relevance vector machine is new to
{maize}! below shows an example of how to use this engine for
regression. currently, the only kernel supported in
maize 0.0.1.9000 is “laplacian”.
additional note, the relevance vector machine may run into a stack overflow error. This typically happens with the RVM when processing large datasets or when the algorithm gets stuck in a recursive loop. cross products are called many times which is computationally intensive due to the training complexity. at this time, may look into an alternative RVM library but not interested in extending to other kernels until C stack issues can be resolved.
# simplified example given training complexity:
x <- seq(-20,20,0.1) |> scale()
y <- sin(x)/x + rnorm(401,sd=0.03) |> scale()
df <- data.frame(x = x, y = y)
train_df <- df |> head(200)
# model params --
rvm_reg_spec <-
rvm_laplace(laplace_sigma = 100) |>
set_mode("regression") |>
set_engine("kernlab")
# fit --
rvm_reg_fit <- rvm_reg_spec |> fit(y ~ ., data = train_df)
# predictions --
preds <- predict(rvm_reg_fit, df |> tail(100))