SVMs
SVMs.RmdExample Overview
corn kernels and svm kernels
say we have three types of corn with different size kernels which create different height corn. we could (1) create a support vector machine that can predict the height of the corn given the kernel type and kernel size, we could also (2) predict the corn type given the kernel size and corn height.
corn kernel dataset
library(parsnip)
library(maize)
library(ggplot2)
# use this later to create a classification field
# -----------------------------------------------
kernel_min <- corn_data$kernel_size |> min()
kernel_max <- corn_data$kernel_size |> max()
kernel_vec <- seq(kernel_min, kernel_max, by = 1)
height_min <- corn_data$height |> min()
height_max <- corn_data$height |> max()
height_vec <- seq(height_min, height_max, by = 1)
corn_grid <- expand.grid(kernel_size = kernel_vec, height = height_vec)
# -----------------------------------------------
maize::corn_data |>
ggplot() +
geom_point(aes(x = kernel_size, y = height, color = type)) +
theme_minimal() +
labs(title = 'corn kernel data') 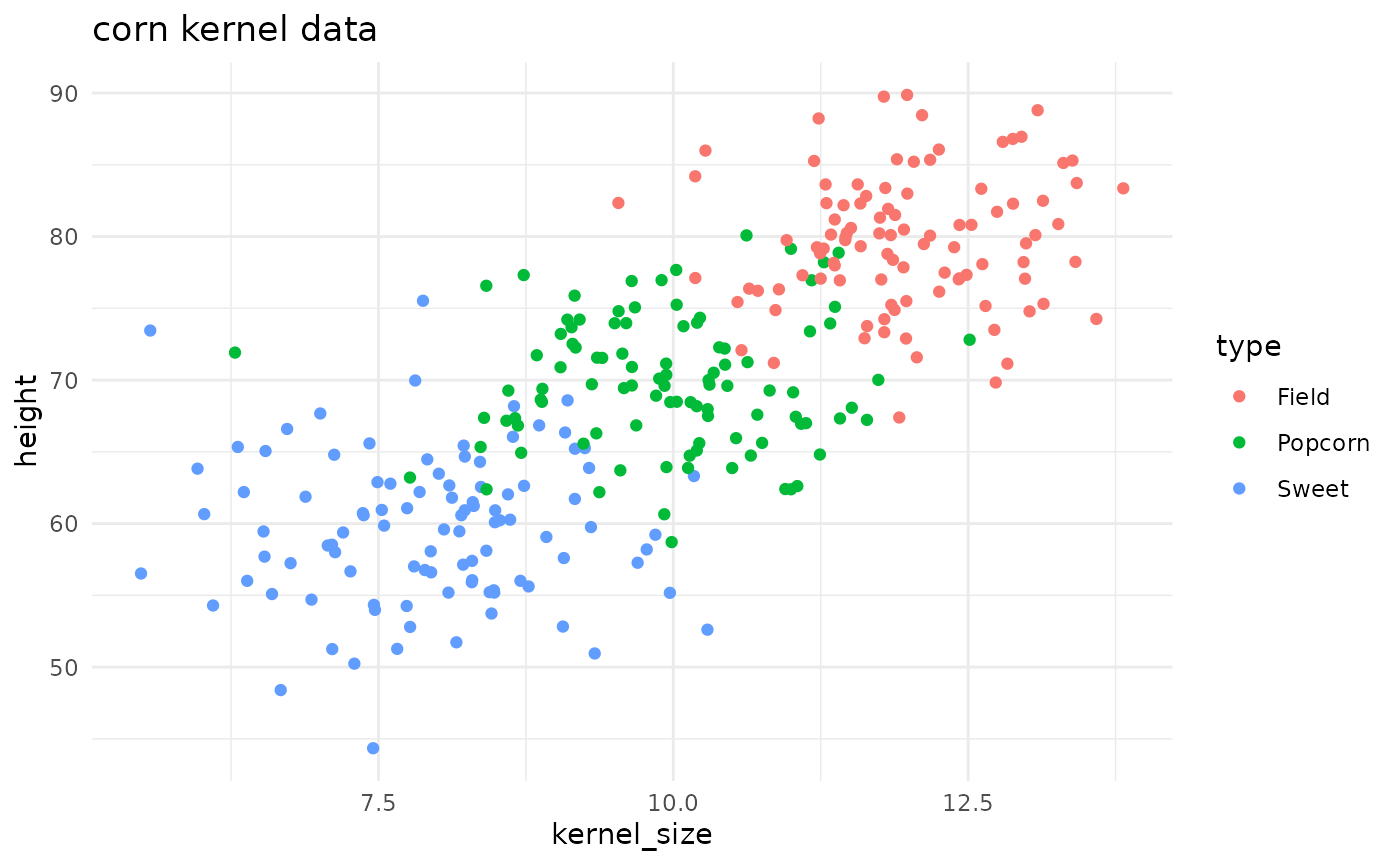
Regression
regression for corn kernel height given a specialty svm kernel
set.seed(31415)
corn_train <- corn_data |> dplyr::sample_frac(.80)
corn_test <- corn_data |> dplyr::anti_join(corn_train)
#> Joining with `by = join_by(height, kernel_size, type)`
# model params --
svm_reg_spec <-
svm_bessel(cost = 1, margin = 0.1) |>
set_mode("regression") |>
set_engine("kernlab")
# fit --
svm_reg_fit <- svm_reg_spec |> fit(height ~ ., data = corn_train)
#> Setting default kernel parameters
# predictions --
preds <- predict(svm_reg_fit, corn_test)
# plot --
corn_test |>
cbind(preds) |>
ggplot() +
geom_point(aes(x = kernel_size, y = height, color = type), shape = 1, size = 2) +
geom_point(aes(x = kernel_size, y = .pred, color = type), shape = 2, size = 3) +
theme_minimal() +
labs(title = 'Bessel Kernel SVM',
subtitle = "corn height prediction") 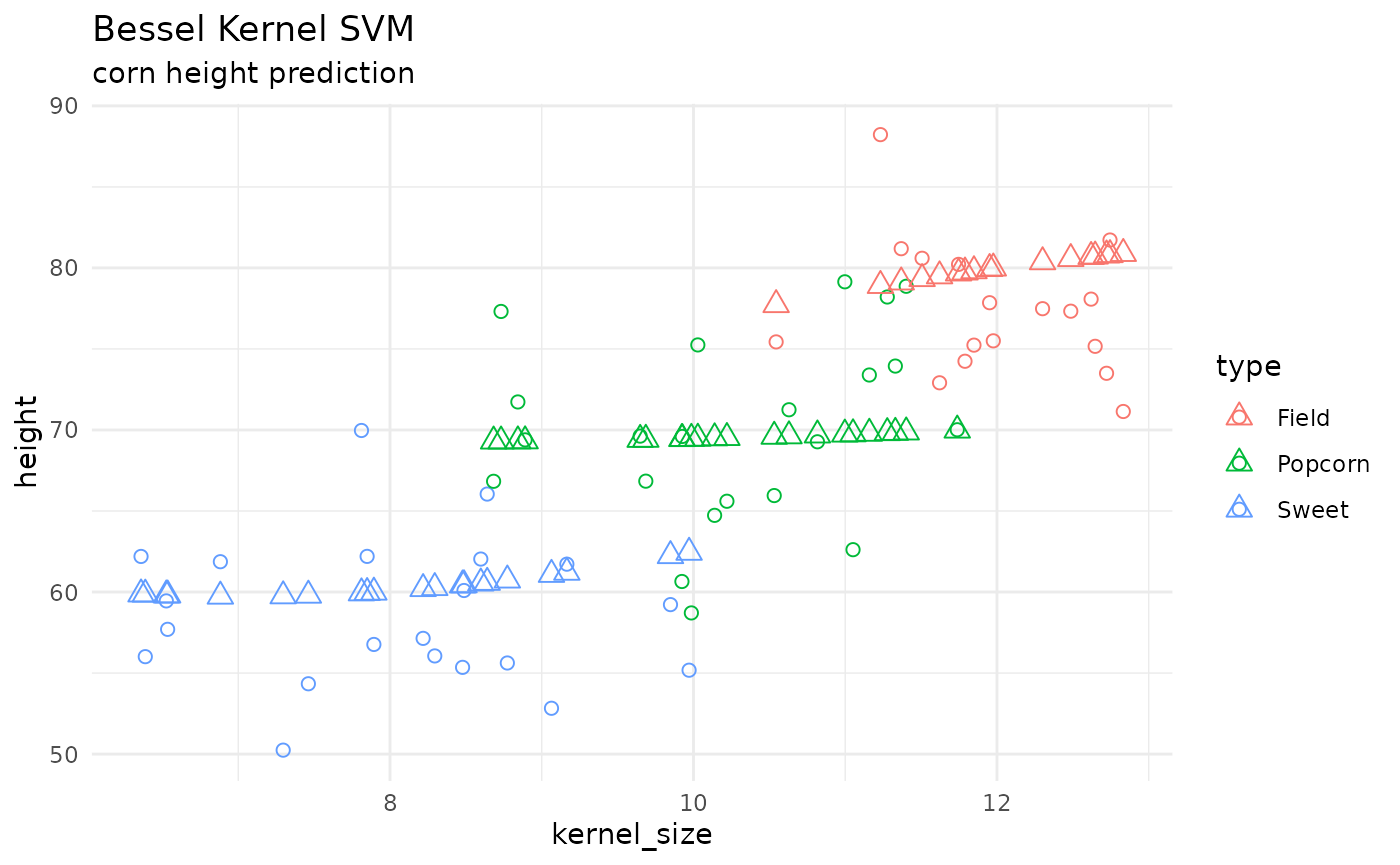
Classification
classification of corn kernel type given a specialty svm kernel
# model params --
svm_cls_spec <-
svm_laplace(cost = 1, margin = 0.1, laplace_sigma = 10) |>
set_mode("classification") |>
set_engine("kernlab")
# fit --
svm_cls_fit <- svm_cls_spec |> fit(type ~ ., data = corn_train)
# predictions --
preds <- predict(svm_cls_fit, corn_grid, "class")
pred_grid <- corn_grid |> cbind(preds)
# plot --
corn_test |>
ggplot() +
geom_tile(inherit.aes = FALSE,
data = pred_grid,
aes(x = kernel_size, y = height, fill = .pred_class),
alpha = .8) +
geom_point(aes(x = kernel_size, y = height, color = type, shape = type), size = 3) +
theme_minimal() +
labs(subtitle = "Laplacian Kernel") +
scale_fill_viridis_d() +
scale_color_manual(values = c("violet", "cyan", "orange"))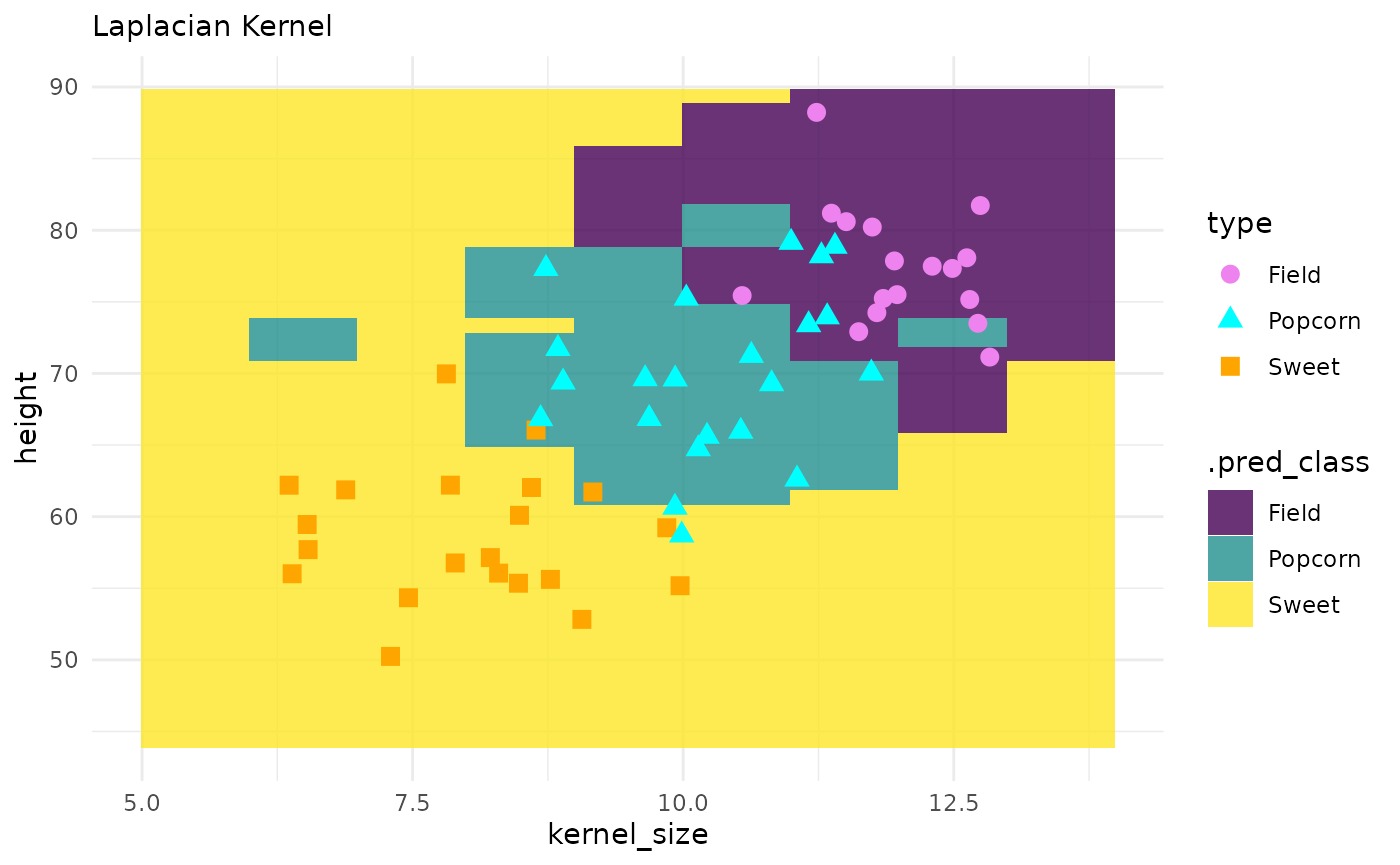
# model params --
svm_cls_spec <-
svm_cossim(cost = 1, margin = 0.1) |>
set_mode("classification") |>
set_engine("kernlab")
# fit --
svm_cls_fit <- svm_cls_spec |> fit(type ~ ., data = corn_train)
# predictions --
preds <- predict(svm_cls_fit, corn_grid, "class")
pred_grid <- corn_grid |> cbind(preds)
# plot --
corn_test |>
ggplot() +
geom_tile(inherit.aes = FALSE,
data = pred_grid,
aes(x = kernel_size, y = height, fill = .pred_class),
alpha = .8) +
geom_point(aes(x = kernel_size, y = height, color = type, shape = type), size = 3) +
theme_minimal() +
labs(subtitle = "Cossim Similarity Kernel") +
scale_fill_viridis_d() +
scale_color_manual(values = c("violet", "cyan", "orange"))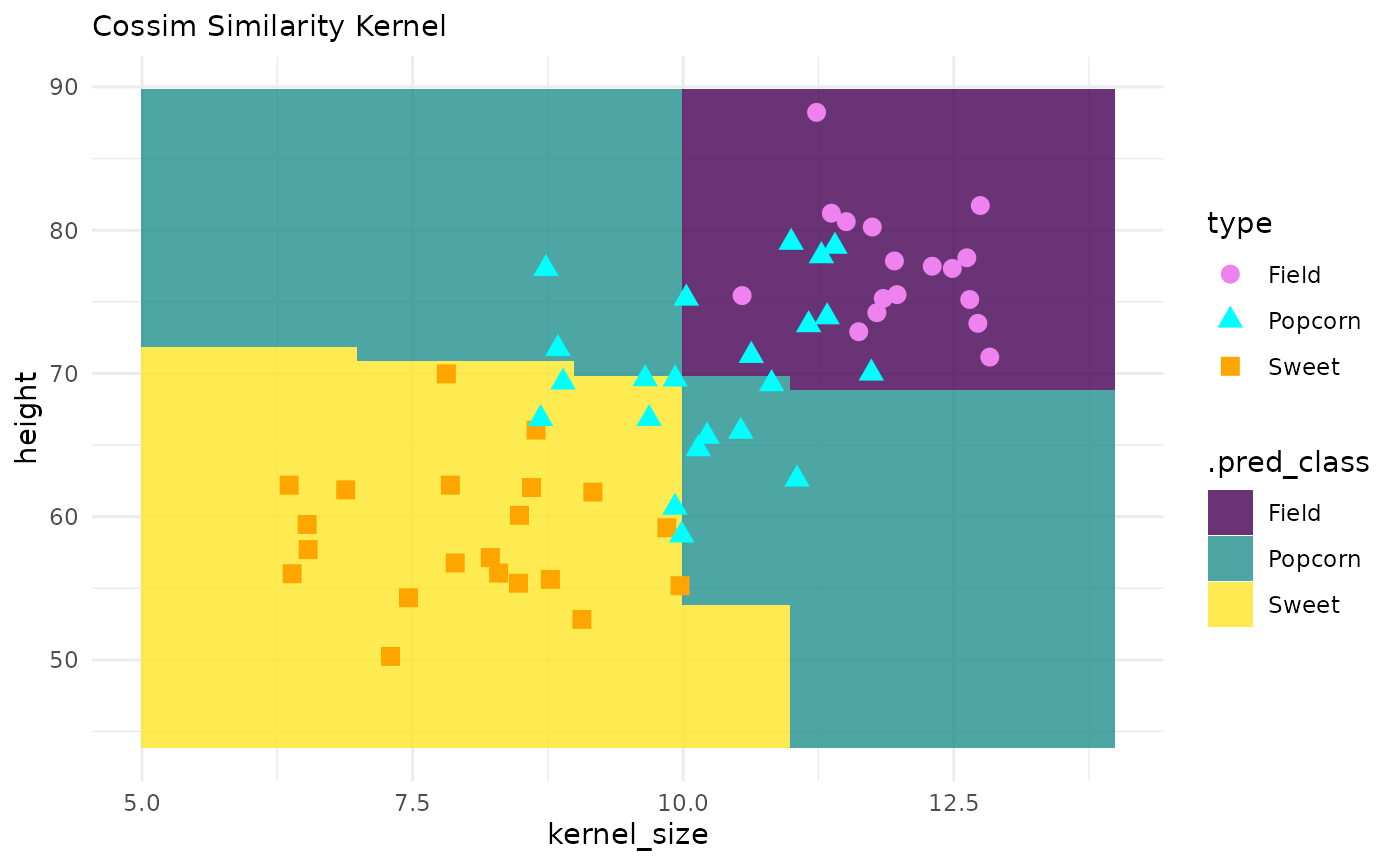
# model params --
svm_cls_spec <-
svm_anova_rbf(cost = 1, margin = 0.1) |>
set_mode("classification") |>
set_engine("kernlab")
# fit --
svm_cls_fit <- svm_cls_spec |> fit(type ~ ., data = corn_train)
#> Setting default kernel parameters
# predictions --
preds <- predict(svm_cls_fit, corn_grid, "class")
pred_grid <- corn_grid |> cbind(preds)
# plot --
corn_test |>
ggplot() +
geom_tile(inherit.aes = FALSE,
data = pred_grid,
aes(x = kernel_size, y = height, fill = .pred_class),
alpha = .8) +
geom_point(aes(x = kernel_size, y = height, color = type, shape = type), size = 3) +
theme_minimal() +
labs(subtitle = "ANOVA RBF Kernel") +
scale_fill_viridis_d() +
scale_color_manual(values = c("violet", "cyan", "orange"))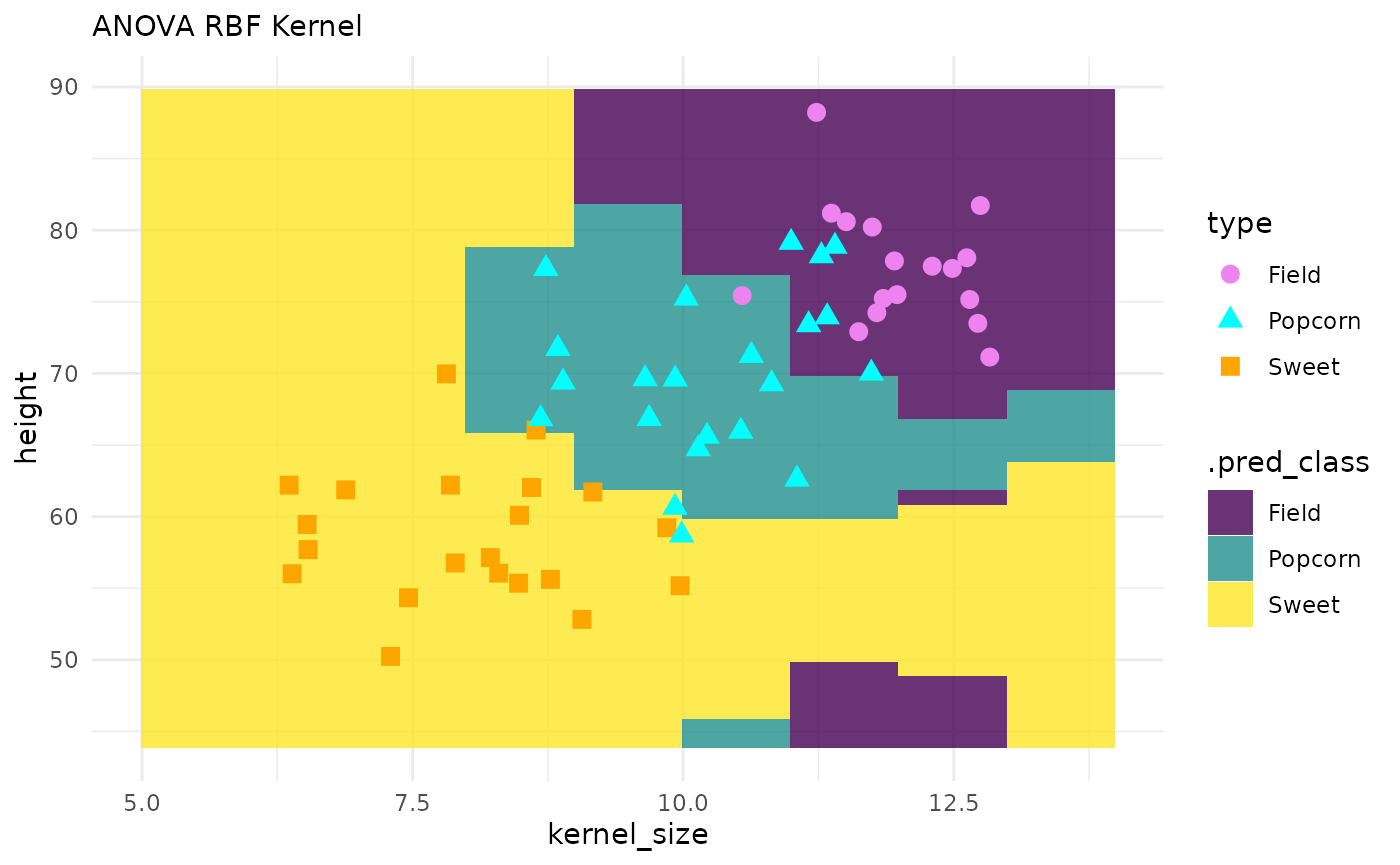
# model params --
svm_cls_spec <-
svm_tanimoto(cost = 1, margin = 0.1) |>
set_mode("classification") |>
set_engine("kernlab")
# fit --
svm_cls_fit <- svm_cls_spec |> fit(type ~ ., data = corn_train)
# predictions --
preds <- predict(svm_cls_fit, corn_grid, "class")
pred_grid <- corn_grid |> cbind(preds)
# plot --
corn_test |>
ggplot() +
geom_tile(inherit.aes = FALSE,
data = pred_grid,
aes(x = kernel_size, y = height, fill = .pred_class),
alpha = .8) +
geom_point(aes(x = kernel_size, y = height, color = type, shape = type), size = 3) +
theme_minimal() +
labs(subtitle = "Tanimoto Kernel") +
scale_fill_viridis_d() +
scale_color_manual(values = c("violet", "cyan", "orange"))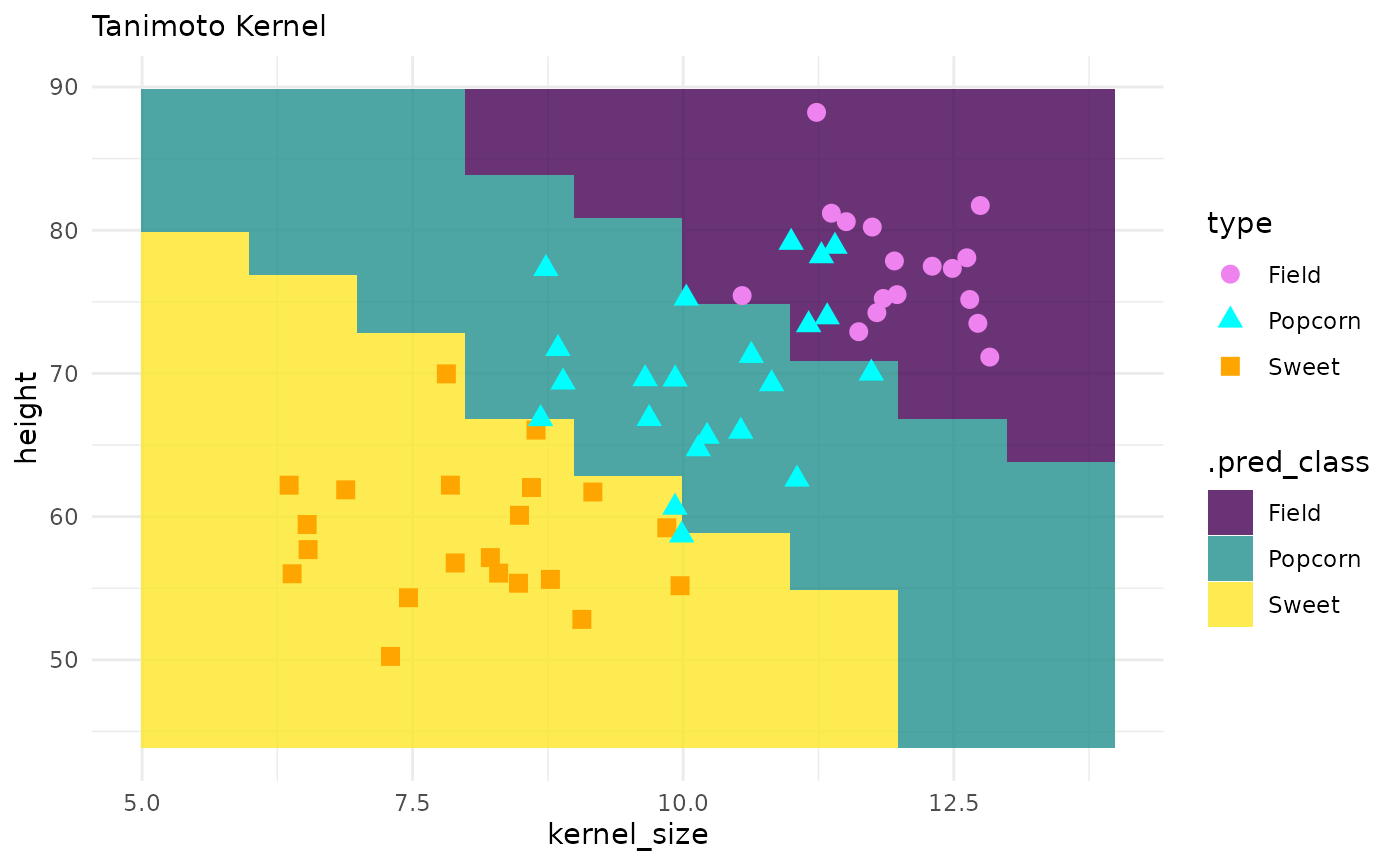
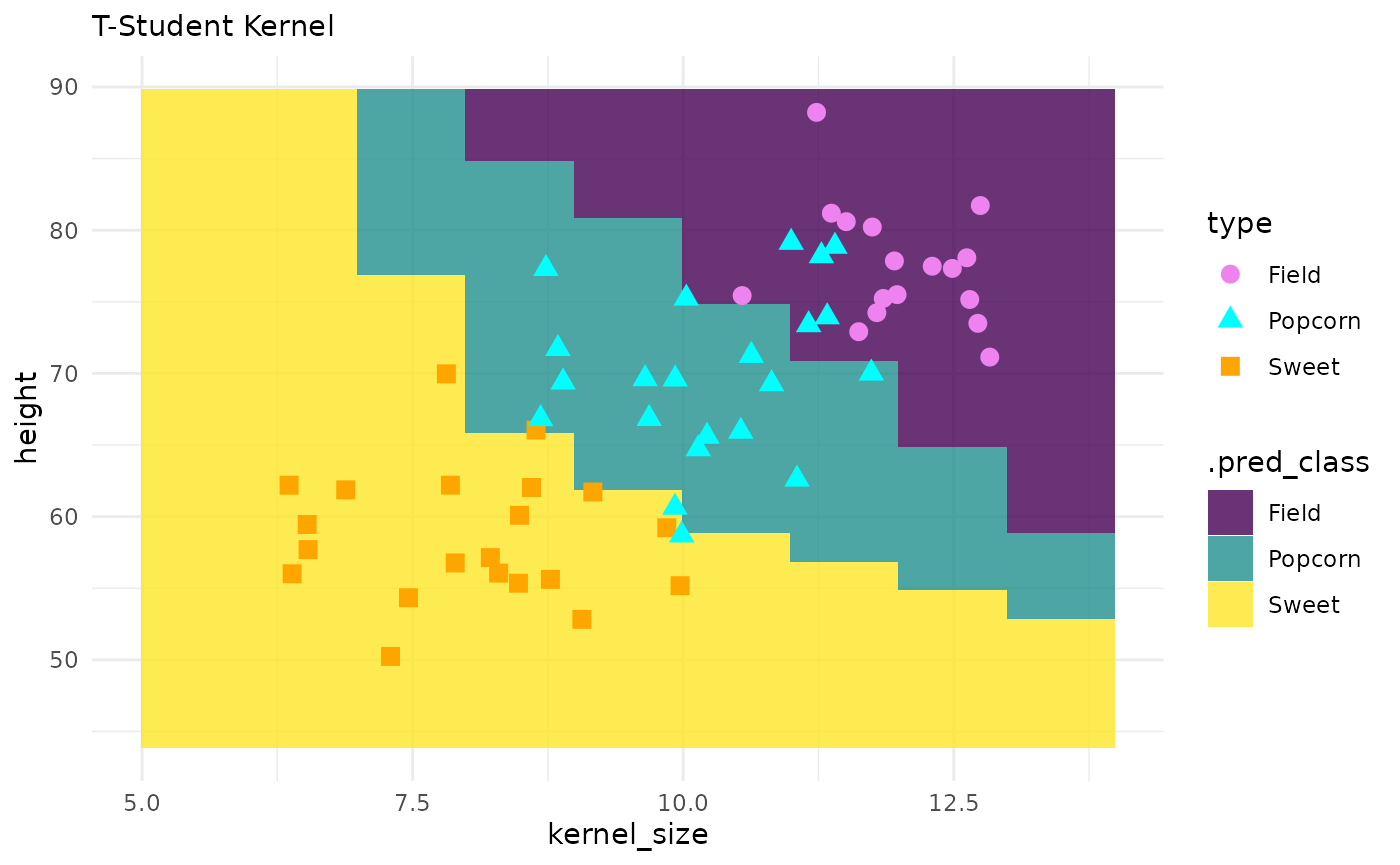
# model params --
svm_cls_spec <-
svm_tstudent(cost = 1, margin = 0.1, degree = 3) |>
set_mode("classification") |>
set_engine("kernlab")
# fit --
svm_cls_fit <- svm_cls_spec |> fit(type ~ ., data = corn_train)
# predictions --
preds <- predict(svm_cls_fit, corn_grid, "class")
pred_grid <- corn_grid |> cbind(preds)
# plot --
corn_test |>
ggplot() +
geom_tile(inherit.aes = FALSE,
data = pred_grid,
aes(x = kernel_size, y = height, fill = .pred_class),
alpha = .8) +
geom_point(aes(x = kernel_size, y = height, color = type, shape = type), size = 3) +
theme_minimal() +
labs(subtitle = "T-Student Kernel") +
scale_fill_viridis_d() +
scale_color_manual(values = c("violet", "cyan", "orange"))