ensembling weak learners in maize
ensembling_weak_learners.Rmd
library(ggplot2)
library(parsnip)
library(maize)
#>
#> Attaching package: 'maize'
#> The following object is masked from 'package:parsnip':
#>
#> check_args
# binary-classification [1,0]
corn_df <- corn_data |>
dplyr::mutate(type = as.character(type)) |>
dplyr::filter(type %in% c("Sweet", "Popcorn")) |>
dplyr::mutate(type = factor(type)) |>
dplyr::mutate(
type = factor(type, levels = c("Sweet", "Popcorn"), labels = c("0", "1"))
)
set.seed(31415)
corn_train <- corn_df |> dplyr::sample_frac(.80) |> as.data.frame()
corn_test <- corn_df |> dplyr::anti_join(corn_train) |> as.data.frame()
#> Joining with `by = join_by(height, kernel_size, type)`
# use this later to create a classification field
# -----------------------------------------------
kernel_min <- corn_df$kernel_size |> min()
kernel_max <- corn_df$kernel_size |> max()
kernel_vec <- seq(kernel_min, kernel_max, by = 1)
height_min <- corn_df$height |> min()
height_max <- corn_df$height |> max()
height_vec <- seq(height_min, height_max, by = 1)
corn_grid <- expand.grid(kernel_size = kernel_vec, height = height_vec)
# -----------------------------------------------Boosting & Bagging with {ebmc} & {maize}
The {ebmc} package supports binary classification
tasks, where the response variable is encoded as [1,
0]. The package provides implementations of several boosting
and bagging methods tailored for imbalanced datasets. A handful of these
have been ported to {maize} including: ebmc::ub(),
ebmc::rus(), and ebmc::adam2(), which are
configured to use e1071::svm() as the base learning
algorithm for ensemble modeling. In {maize}, the current kernel
supported is a radial basis function.
Extending {baguette}’s implementation for SVM’s may be added at a later date but note the backend is currently {ebmc} and the technique differs.
Random Under Sampling (RUS) Bagging with SVMs
# model params --
bagged_svm_spec <-
bag_svm_rbf(num_learners = 50,
imb_ratio = 1) |>
set_mode("classification") |>
set_engine("ebmc")
# fit --
bag_svm_class_fit <- bagged_svm_spec |> fit(type ~ ., data = corn_train)
# predictions --
preds <- predict(bag_svm_class_fit, corn_grid, "class")
pred_grid <- corn_grid |> cbind(preds)
# plot --
corn_test |>
ggplot() +
geom_tile(inherit.aes = FALSE,
data = pred_grid,
aes(x = kernel_size, y = height, fill = .pred_class),
alpha = .8) +
geom_point(aes(x = kernel_size, y = height, color = type, shape = type), size = 3) +
theme_minimal() +
labs(title = "RUS Bagging with RBF SVMs",
subtitle = "corn class prediction") +
scale_fill_viridis_d() +
scale_color_manual(values = c("violet", "orange"))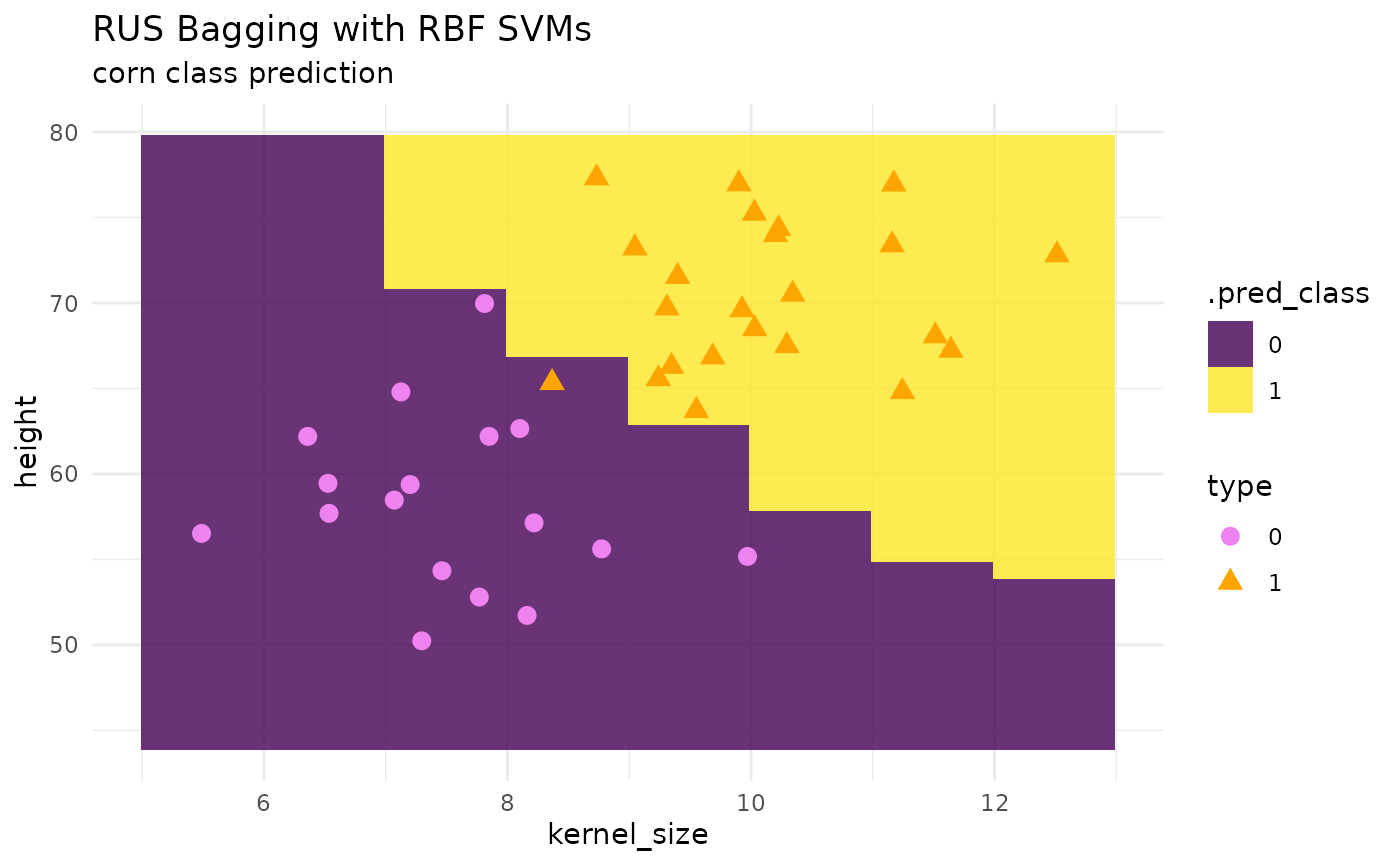
Random Under Sampling with Boosting, RUSBoost with SVMs
# model params --
boost_svm_spec <-
rus_boost_svm_rbf(num_learners = 50,
imb_ratio = 1) |>
set_mode("classification") |>
set_engine("ebmc")
# fit --
boost_svm_class_fit <- boost_svm_spec |> fit(type ~ ., data = corn_train)
# predictions --
preds <- predict(boost_svm_class_fit, corn_grid, "class")
pred_grid <- corn_grid |> cbind(preds)
# plot --
corn_test |>
ggplot() +
geom_tile(inherit.aes = FALSE,
data = pred_grid,
aes(x = kernel_size, y = height, fill = .pred_class),
alpha = .8) +
geom_point(aes(x = kernel_size, y = height, color = type, shape = type), size = 3) +
theme_minimal() +
labs(title = "RUS Boosting with RBF SVMs",
subtitle = "corn class prediction") +
scale_fill_viridis_d() +
scale_color_manual(values = c("violet", "orange"))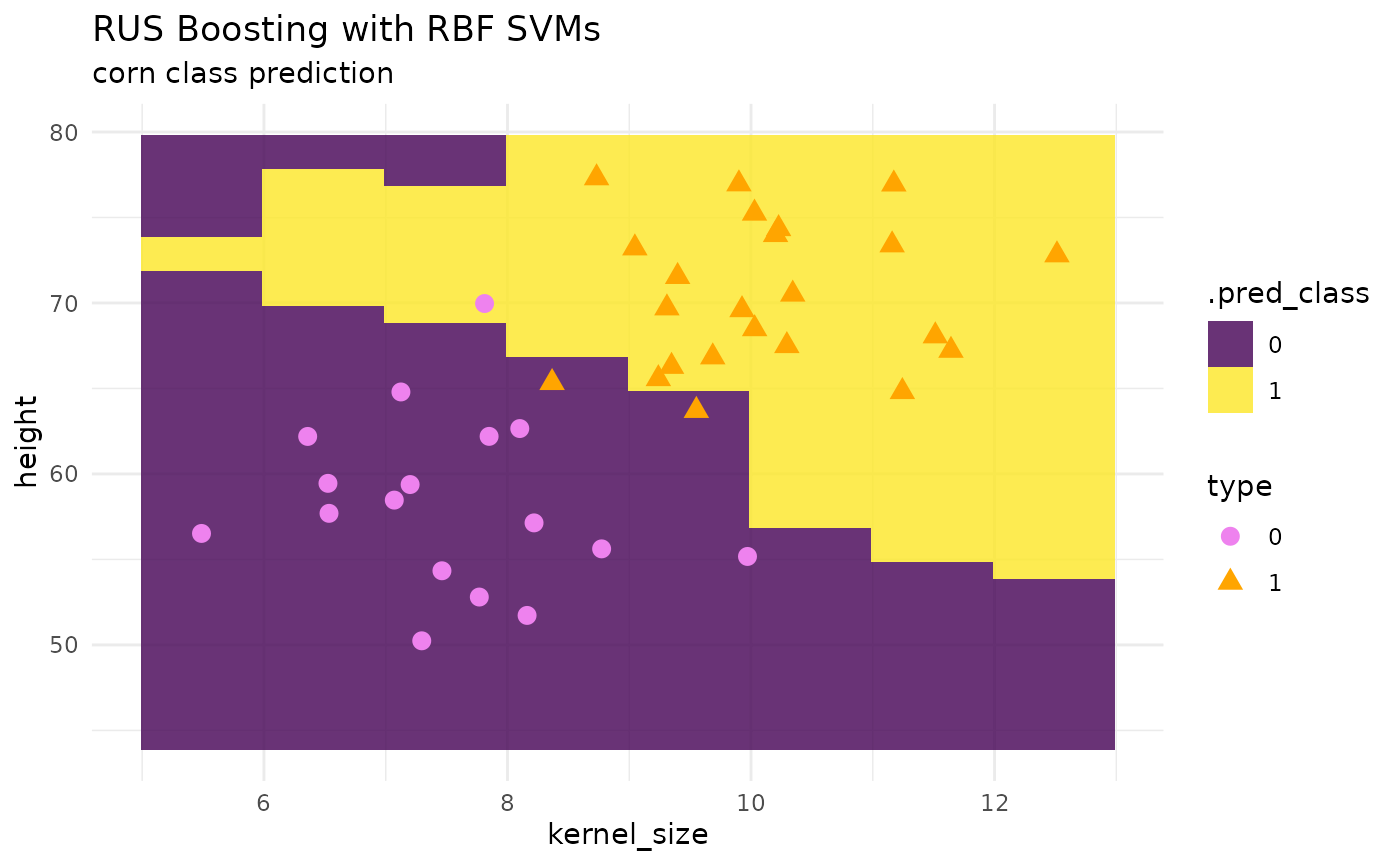
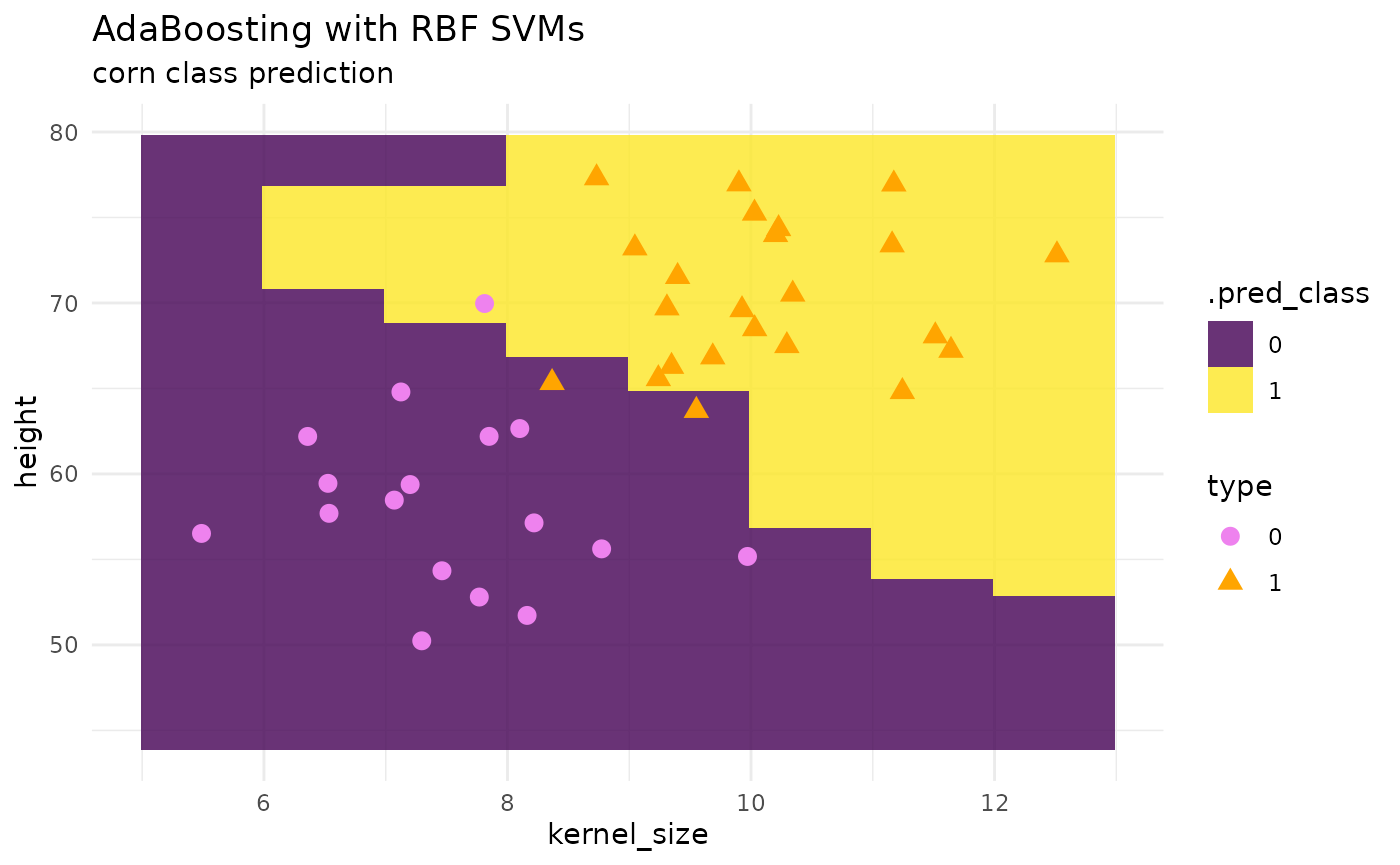
Adaptive Boosting, AdaBoost with SVMs
# model params --
adaboost_svm_spec <-
ada_boost_svm_rbf(num_learners = 50,
imb_ratio = 1) |>
set_mode("classification") |>
set_engine("ebmc")
# fit --
adaboost_svm_class_fit <- adaboost_svm_spec |> fit(type ~ ., data = corn_train)
# predictions --
preds <- predict(adaboost_svm_class_fit, corn_grid, "class")
pred_grid <- corn_grid |> cbind(preds)
# plot --
corn_test |>
ggplot() +
geom_tile(inherit.aes = FALSE,
data = pred_grid,
aes(x = kernel_size, y = height, fill = .pred_class),
alpha = .8) +
geom_point(aes(x = kernel_size, y = height, color = type, shape = type), size = 3) +
theme_minimal() +
labs(title = "AdaBoosting with RBF SVMs",
subtitle = "corn class prediction") +
scale_fill_viridis_d() +
scale_color_manual(values = c("violet", "orange"))